FUN with Legumes and Zinc — But Is More Nitrogen Always Better?
Legumes like beans, peas and lentils are unique among crops for their ability to interact with soil bacteria to convert or “fix” nitrogen into a usable form of nutrients. However, this energy-intensive biological process is reduced when nitrogen is already abundant in the soil, either through natural processes or through the application of synthetic fertiliser.
Researchers have recently discovered the genetic regulator that turns off nitrogen fixation when soil nitrate levels are high. As part of the experiment, they also removed the gene in model legumes, ensuring they continued to fix nitrogen regardless of the soil environment. The study, which is currently funded by Bill & Melinda Gates Agricultural Innovations, were published in Nature.
According to the researchers, increasing the biological ability of legumes to fix nitrogen could help increase crop growth and yield while also reducing the need for synthetic fertilizers, which contribute to agriculture’s environmental footprint.
“From an agricultural perspective, continued nitrogen fixation could be a beneficial trait that increases nitrogen availability, both for the legume and for future crops that rely on the nitrogen left behind in the soil after legumes are grown,” said lead author Dr. Dugald Reid.
The team discovered the regulator known as “Fixation Under Nitrate” (FUN) after screening 150,000 individual legume plants in which genes had been knocked out to identify how plants control the switch from nitrogen fixation to soil nitrogen uptake. FUN, which is a type of gene known as a transcription factor and controls the levels of other genes, was found to be present in legumes regardless of whether it was active or inactive, and irrespective of nitrogen levels.
The team then used a combination of biochemistry, gene expression studies and microscopy to find that FUN forms into long protein filaments when it is inactive. This led to a secondary discovery — that zinc levels play a role in triggering FUN to become active and shut down nitrogen fixation. Lower zinc concentrations in the nodule, which occur in response to higher levels of soil nitrate, activate FUN. FUN then directly targets multiple pathways to initiate breakdown of the nodule.
“We found that changing soil nitrogen alters the levels of zinc in the plant. Zinc had not previously been linked to the regulation of nitrogen fixation, but our study found that a change in zinc levels in turn activates FUN, which then controls a large number of genes that shut down nitrogen fixation,” said co-author Dr. Kasper Andersen. “Removing FUN therefore creates a condition in which nitrogen fixation is no longer shut down by the plant.”
| Excess nitrogen — from either fertilizer or rhizobium — could be a costly mistake While obtaining more nitrogen from legumes in order to reduce the application of synthetic nitrogen fertilizers sounds, on the face of it, like a good thing, a word of caution is still in order. That is, if plants (legumes aren’t the only plants that harbor bacteria that can fix atmospheric nitrogen) are artificially stimulated to produce excess nitrogen — more than the plant and/or the soil need — that extra nitrogen will begin to consume whatever carbon is in the soil. Less carbon in the soil means poor aggregation, which means compaction. Well-intending farmers who include too many legumes in their cover crop mixes figure this out. Plants need the right amount of nitrogen (and of zinc, cobalt and every other mineral). Too much can be just as bad as too little. |
Purple Marine Bacteria Makes an Excellent Eco-Friendly Fertilizer
The ecologically harmful side effects of overusing common inorganic nitrogen fertilizers — substantial greenhouse gas emissions, contaminated ground water and poor soil quality — are increasingly being recognized. Yet organic fertilizers such as compost and manure can lead to excessively salty soil and overabundance of some minerals.
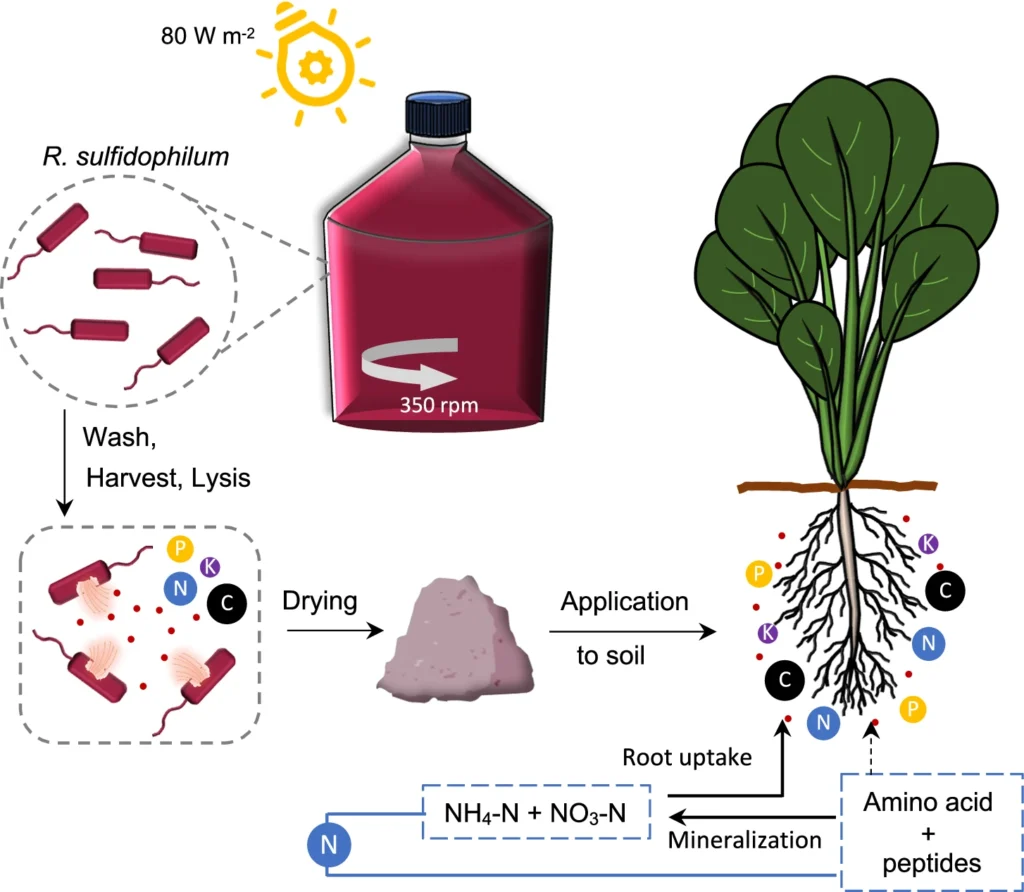
Researchers at the Japanese basic and applied science organization RIKEN recently tested purple non-sulfur bacteria as a potential nitrogen fertilizer source. PNSBs are known to have enzymes that help them take nitrogen from the atmosphere and incorporate it into proteins. The team mashed up the PNSB R. sulfidophilum and generated dried biomass from the released cellular material. Analysis showed that the nitrogen content of the PNSB fertilizer was 11 percent by weight — much higher than what is found in other organic fertilizers, including biomass made from other microbes or microalgae.
The researchers compared how well the plant komatsuna (Brassica rapa var. perviridis) grew when assisted by either inorganic fertilizers or the new PNSB biomass fertilizer. Experiments showed that the biomass fertilizer boosted plant growth just as well as nitrogen-rich inorganic fertilizers, at both cool and warm temperatures. Even when the PNSB fertilizer contained up to four times the amount of nitrogen, soil pH and salinity remained normal.
The PNSB biomass fertilizer has a low carbon-to-nitrogen ratio, and the nitrogen is released for plant use relatively slowly compared with inorganic fertilizers — about 60 percent in 30 days. This means fewer carbon dioxide and nitrous oxide emissions and less nitrogen leaching.
| Quick take: Why are farmers moving toward regenerative systems? From “Relationships of regeneration in Great Plains commodity agriculture,” recently published in Agriculture and Human Values: “A decades-old regenerative agriculture movement is growing rapidly, but not due to the incentives offered by companies’ carbon programs. On the contrary, farmers are adopting regenerative practices to cut their dependence on corporate agrochemical inputs and expertise, and to thereby achieve technology sovereignty…. These findings provide insight into farmers’ skepticism of private-sector carbon farming programs and highlight the value of attention to the multiple types of relationship change that accompany and facilitate regenerative transitions.” |
Mycoviruses Enhance Fungicide Effectiveness against Plant Pathogens
As detrimental as viruses may sound, they can be helping hands for farmers when it comes to dealing with plant pathogens.
Osaka Metropolitan University scientists have discovered that a mycovirus that infects the plant pathogenic oomycete Globisporangium ultimum can increase the latter’s sensitivity to specific fungicides. Their findings could lead to innovative approaches for controlling plant diseases, reducing reliance on chemical treatments and minimizing agricultural loss. The results were published in Microbiological Research.
Mycoviruses, or fungal viruses, infect fungi as well as fungus-like organisms such as oomycetes. More commonly known as water molds, oomycetes include some of the most devastating plant pathogens and pose severe threats to global food security. When these oomycetes are infected by certain mycoviruses, however, their ability to cause disease can be weakened — a phenomenon known as hypovirulence — making mycoviruses potential biocontrol agents.
Mycoviral infections are multifaceted; they can reduce or increase virulence or remain hidden without obvious symptoms. Despite the growing number of mycoviruses identified recently, their effects on host oomycetes have remained largely unexplored.
The research team zeroed in on G. ultimum, a major soil-borne oomycete responsible for damping-off and root rot in many plant species. They first created a virus-free isogenic strain of G. ultimum through high-temperature cultivation and then compared its characteristics and gene expression to the virus-infected isogenic strain.
The results show that compared to the virus-free isogenic strain, the virus-infected isogenic strain was more sensitive to metalaxyl, one of the four tested fungicides. No significant differences in growth rate and structure were observed between these isogenic strains in the absence of metalaxyl. Using a high-throughput screening technique called RNA-seq to analyze gene expression profile, the researchers found that the virus-infected isogenic strain had lower expression of certain genes known as ABC-type transporters, which are known to contribute to fungicide resistance.
These findings advance the current understanding of the roles that mycoviruses play and their potential for sustainable agriculture. The team plans to further explore the promise of mycoviruses as a biological control tool across different species and environmental conditions.
| Demystifying biological plant protection products The new BioProtection Portal (bioprotectionportal.com) aims to make finding appropriate biological pest protection resources easy. Just enter your region, crop and pest, and you’ll receive a listing of potentially applicable products that are more natural and less ecologically damaging than the typical synthetic inputs. |
| The Right Goal, the Wrong Means A pair of new studies aim to help do the right thing — increase total photosynthesis — albeit via unnatural means that will deliver unknown side effects. Ecological and regenerative farmers already know how to maximize photosynthesis — and feed the world — without these forms of hubristic human “progress.” Fine-Tuning Leaf Angle with CRISPR Improves Sugarcane Yield Sugarcane is the world’s largest crop by biomass yield, producing 80 percent of sugar and 40 percent of biofuel worldwide. The plant’s size and efficient use of water and light make it a prime candidate to produce advanced renewable, value-added bioproducts and biofuels. However, as a hybrid of Saccharum officinarum and Saccharum spontaneum, sugarcane has the most complex genome of all crops. This complexity means that improving sugarcane through conventional breeding is challenging. Because of this, researchers at the University of Florida recently turned to a gene editing tool —CRISPR/Cas9 — to precisely target the sugarcane genome for improvement. These genetic tweaks allowed the sugarcane to capture more sunlight, which in turn increased the amount of biomass produced. Their results were published in Plant Biotechnology Journal. The sugarcane genome’s complexity is due in part to its high levels of redundancy: It possesses many copies of each gene. The phenotype that a sugarcane plant displays, therefore, typically depends on the cumulative expression of the multiple copies of a certain gene. This study focused on LIGULELESS1, or LG1, a gene that plays a major role in determining leaf angle in sugarcane. Leaf angle, in turn, determines how much light can be captured by the plant, which is critical for biomass production. Since sugarcane’s highly redundant genome contains 40 copies of LG1, the researchers were able to fine-tune the leaf angle by editing different numbers of copies of this gene, resulting in slightly different leaf angles depending on how many copies of LG1 were edited. When the researchers grew sugarcane in field trials, they found that the upright leaf phenotypes allowed more light to penetrate the canopy, which resulted in increased biomass yield. One sugarcane line in particular, which contained edits in about 12 percent of the LG1 copies and showed a 56 percent decrease in leaf inclination angle, had an 18 percent increase in dry biomass yield. By optimizing sugarcane to capture more light, these gene edits increase biomass yield without having to add more fertilizer to the fields. Changes Upstream: Researchers Use CRISPR/Cas9 to Increase Gene Expression and Alter Photosynthesis A team from the University of California, Berkeley has produced an increase in gene expression in a food crop by changing its upstream regulatory DNA. While other studies have used CRISPR/Cas9 gene-editing to knock out or decrease the expression of genes, this new research, published in Science Advances, is the first unbiased gene-editing approach to increase gene expression and downstream photosynthetic activity. “Tools like CRISPR/Cas9 are accelerating our ability to fine-tune gene expression in crops, rather than just knocking out genes or turning them ‘off,’” said lead author Dhruv Patel-Tupper. “Past research has shown that this tool can be used to decrease expression of genes involved in important trade-offs, such as those between plant architecture and fruit size. This is the first study, to our knowledge, where we asked if we can use the same approach to increase the expression of a gene and improve downstream activity in an unbiased way. ”Unlike synthetic biology strategies that use genes from other organisms to improve photosynthesis, the genes involved in the photoprotection process are naturally found in all plants. Inspired by a 2018 study that improved the water-use efficiency of a model crop by overexpressing one of these genes, PsbS, in plants, the lab wanted to figure out how to change the expression of a plants’ native genes without adding foreign DNA. According to the Food and Agriculture Organization, rice supplies at least 20 percent of the world’s calories, and because it has only one copy of each of the three key photoprotection genes in plants, it was an ideal model system for this gene editing study. The lab’s plan was to use CRISPR/Cas9 to change the DNA upstream of the target gene, which controls how much of the gene is expressed and when. They wondered if making those changes would have an impact on downstream activity and by how much. “The changes in the DNA that increased gene expression were much bigger than we expected and bigger than we’ve really seen reported in other similar stories,” said Patel-Tupper. “We were a little bit surprised, but I think it goes to show how much plasticity plants and crops have. They’re used to these big changes in their DNA from millions of years of evolution and thousands of years of domestication. As plant biologists, we can leverage that ‘wiggle room’ to make large changes in just a handful of years to help plants grow more efficiently or adapt to climate change. ”In this study, researchers learned that inversions, or “flipping” of the regulatory DNA, resulted in increased gene expression of PsbS. Unique to this project, after the largest inversion was made to the DNA, the team members conducted an RNA sequencing experiment to compare how the activity of all genes in the rice genome changed with and without their modifications. What they found was a very small number of differentially expressed genes, much smaller than similar transcriptome studies, suggesting their approach did not compromise the activity of other essential processes. Patel-Tupper added that while the team showed that this method is possible, it’s still relatively rare. Around 1 percent of the plants they generated had the desired phenotype. “We showed a proof-of-concept here, that we can use CRISPR/Cas9 to generate variants in key crop genes and get the same leaps as we would in traditional plant breeding approaches, but on a very focused trait that we want to engineer and at a much faster timescale,” said Patel-Tupper. “It’s definitely more difficult than using a transgenic plant approach, but by changing something that is already there, we may be able to preempt regulatory issues that can slow how quickly we get tools like this into the hands of farmers.” |














